Explore
#metabolic #networks!
Join our online course “Constraint-based modeling & design with CellNetAnalyzer & CNApy” 🧬
🗓 4–5 Nov 25
🔎 Theory, live demos & hands-on sessions—no prereqs!
#Beginner course!
🔗 More Information and Registration:
https://www.denbi.de/training-courses-2025/1912-constraint-based-modeling-and-design-of-metabolic-networks-with-cellnetanalyzer-and-cnapy-2#SystemsBiology #FluxBalance
Virtual cells
ideas, work and experiments
🔎 How can AI-powered tools help scientists uncover hidden connections in disease research?
🔗 Darling (v2.0): Mining disease-related databases for the detection of biomedical entity associations. Computational and Structural Biotechnology Journal, DOI: https://doi.org/10.1016/j.csbj.2025.06.025
📚 CSBJ: https://www.csbj.org/
#AIinScience #BiomedicalTextMining #Genomics #PrecisionMedicine #Bioinformatics #OpenScience #BiomedicalResearch #BiomedicalAI #LiteratureMining #SystemsBiology #Omics #KnowledgeDiscovery
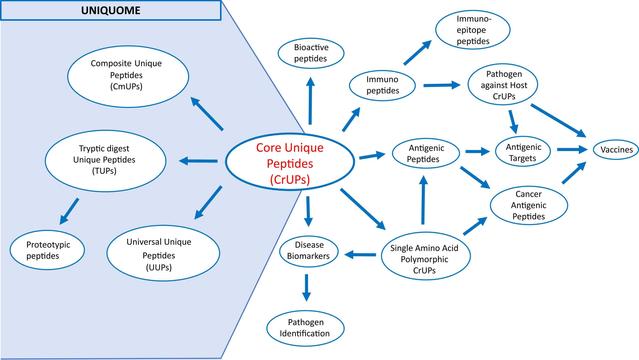
🧬 Are we overcomplicating proteomics—when a few unique sequences can tell the whole story?
🔗 Uniquome: Construction and decoding of a novel proteomic atlas that contains new peptide entities. Computational and Structural Biotechnology Journal, DOI: https://doi.org/10.1016/j.csbj.2025.05.027
📚 CSBJ: https://www.csbj.org/
#Proteomics #Uniquome #PeptideAtlas #Bioinformatics #PrecisionBiology #TranslationalResearch #SystemsBiology #MolecularBiology #PrecisionMedicine
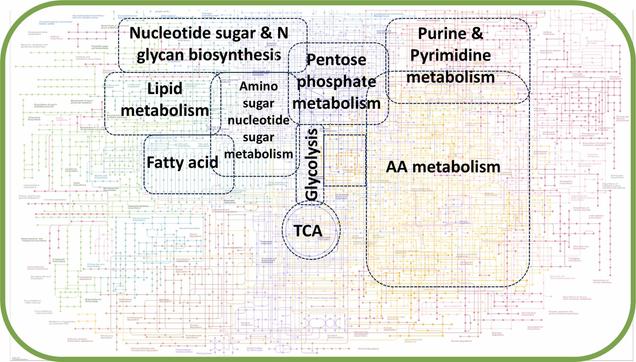
Our (@kaletalab) new study uncovers multi-level metabolic disruptions in #IBD involving reduced #HostMicrobiomeInteractions.
Elevated tryptophan catabolism → NAD biosynthesis impaired.
Disrupted nitrogen homeostasis affects polyamine/glutathione metabolism.
Suppressed C1 cycle alters NAD/phospholipid profiles.
#MetabolicModeling predicts dietary interventions to restore #Microbiome balance.
Read and share: https://rdcu.be/eo7B3
#SystemsBiology #Microbiome #MultiOmics #CAU #UKSH #Kiel
🔬Lennox: Math Abstraction vs. Empirical Reality, how come a mathematician can defend Christianity?
#systemsbiology #atheism #christianity https://www.youtube.com/watch?v=y3lADKLRRa4🔬Lennox: Math Abstraction vs. Empirical Reality, how come a mathematician can defend Christianity?

John Lennox and Systems Biology: A Misuse of Complexity? (Alex O’Connor)
Lets talk #science for a change. We submitted a pre-print of the software I'm working on for my PhD thesis! Beautifully named manvr3d (aka, Manfred), we combine conventional #CellTracking software with #VirtualReality and a deep learning model to create a super fast human-in-the-loop system for tracking cells in 3D through time.
https://arxiv.org/abs/2505.03440
#Paper #ComputerScience #Biology #SystemsBiology
1/5

manvr3d: A Platform for Human-in-the-loop Cell Tracking in Virtual Reality
We propose manvr3d, a novel VR-ready platform for interactive human-in-the-loop cell tracking. We utilize VR controllers and eye-tracking hardware to facilitate rapid ground truth generation and proofreading for deep learning-based cell tracking models. Life scientists reconstruct the developmental history of organisms on the cellular level by analyzing 3D time-lapse microscopy images acquired at high spatio-temporal resolution. The reconstruction of such cell lineage trees traditionally involves tracking individual cells through all recorded time points, manually annotating their positions, and then linking them over time to create complete trajectories. Deep learning-based algorithms accelerate this process, yet depend heavily on manually-annotated high-quality ground truth data and curation. Visual representation of the image data in this process still relies primarily on 2D renderings, which greatly limits spatial understanding and navigation. In this work, we bridge the gap between deep learning-based cell tracking software and 3D/VR visualization to create a human-in-the-loop cell tracking system. We lift the incremental annotation, training and proofreading loop of the deep learning model into the 3rd dimension and apply natural user interfaces like hand gestures and eye tracking to accelerate the cell tracking workflow for life scientists.
🚨 New paper out!
Novel algorithms for uncertainty quantification in systems biology using conformal inference.
Joint work with Alberto Portela and Marcos Matabuena.
📄 Read more: https://doi.org/10.1371/journal.pcbi.1013098
#SystemsBiology #UQ

Conformal prediction for uncertainty quantification in dynamic biological systems
Author summary Uncertainty quantification involves determining how confident we are in the predictions made by mathematical models. This process is vital in the field of systems biology because it helps us understand and predict how these systems behave, despite their complexity. Typically, Bayesian statistics are used for this task. Although powerful, these methods often require specific prior information and make assumptions that may not always hold true for biological systems. Additionally, they struggle when we have limited data, and can be slow for large models. To address these issues, here we have developed two new algorithms based on conformal inference methods. These algorithms offer excellent reliability and scalability. Testing in various scenarios has demonstrated that they outperform traditional Bayesian methods, particularly when applied to large models. Our approach provides a new, general, and flexible method for quantifying uncertainty in dynamic biological models.