🚨 New paper out!
Novel algorithms for uncertainty quantification in systems biology using conformal inference.
Joint work with Alberto Portela and Marcos Matabuena.
📄 Read more: https://doi.org/10.1371/journal.pcbi.1013098
#SystemsBiology #UQ
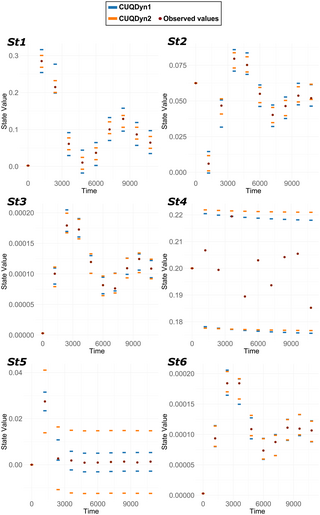
Conformal prediction for uncertainty quantification in dynamic biological systems
Author summary Uncertainty quantification involves determining how confident we are in the predictions made by mathematical models. This process is vital in the field of systems biology because it helps us understand and predict how these systems behave, despite their complexity. Typically, Bayesian statistics are used for this task. Although powerful, these methods often require specific prior information and make assumptions that may not always hold true for biological systems. Additionally, they struggle when we have limited data, and can be slow for large models. To address these issues, here we have developed two new algorithms based on conformal inference methods. These algorithms offer excellent reliability and scalability. Testing in various scenarios has demonstrated that they outperform traditional Bayesian methods, particularly when applied to large models. Our approach provides a new, general, and flexible method for quantifying uncertainty in dynamic biological models.