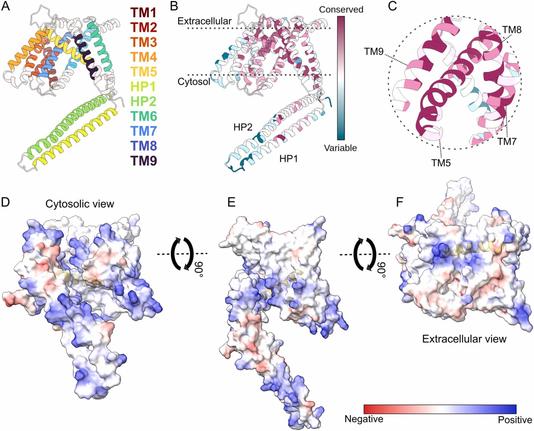
🧩 What if the diverse functions of a single protein all trace back to one structural superfamily?
🔗 Evolutionary and structural bioinformatics identifies GPR89 as a conserved member of the LIMR protein superfamily. Computational and Structural Biotechnology Journal, DOI: https://doi.org/10.1016/j.csbj.2025.11.003
📚 CSBJ: https://www.csbj.org/
#StructuralBiology #ComputationalBiology #MolecularEvolution #ProteinScience #Genomics #Proteomics #AlphaFold #MembraneProteins #Genetics #Bioinformatics
💪 Could one molecular “typo” break the balance between strength and flexibility in our muscles?
🔗 Dynamical features of smooth muscle actin pathological mutants: The arginine-257(258)-Cysteine cases. Computational and Structural Biotechnology Journal, DOI: https://doi.org/10.1016/j.csbj.2025.02.010
📚 CSBJ: https://www.csbj.org/
#ComputationalBiology #MolecularDynamics #ProteinScience #Actin #RareDiseases #PrecisionMedicine #Biophysics #CryoEM #Bioinformatics #DrugDiscovery #VisceralMyopathy #AorticAneurysm
🧬 Can we turn evolutionary signatures into maps of protein function?
🔗 Analysis of Fbox substrate adapter proteins using ProteoSync, a program for projection of evolutionary conservation onto protein atomic coordinates. Computational and Structural Biotechnology Journal, DOI: https://doi.org/10.1016/j.csbj.2025.09.012
📚 CSBJ: https://www.csbj.org/
#ProteinScience #StructuralBiology #ProteoSync #ComputationalBiology #Bioinformatics #EvolutionaryBiology #MolecularBiology #PyMOL
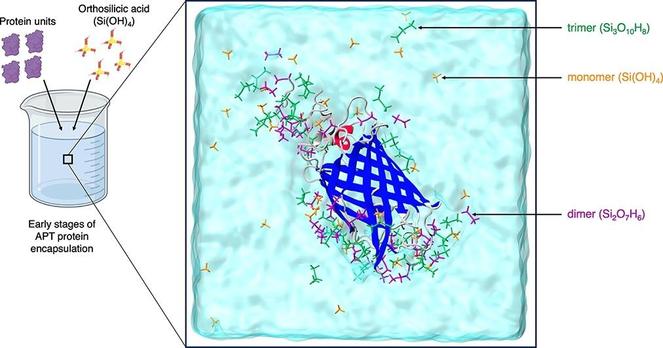
🪨 Can sand-like molecules help us map the secrets of protein structure?
🔗 Adsorption of silica oligomers on biomolecules: Structural and dynamical insights for atom probe tomography via classic molecular dynamics simulations. Computational and Structural Biotechnology Journal, DOI: https://doi.org/10.1016/j.csbj.2025.06.004
📚 CSBJ: https://www.csbj.org/
#AtomProbeTomography #MolecularDynamics #Biophysics #ProteinStructure #Nanotech #ComputationalBiology #StructuralBiology #Biomaterials #ProteinScience
⚛️ Have proteins evolved to operate at a quantum sweet spot between conduction and insulation?
🔗 Holographic nature of critical quantum states of proteins. Computational and Structural Biotechnology Journal, DOI: https://doi.org/10.1016/j.csbj.2025.05.049
📚 CSBJ Quantum Biology and Biophotonics: https://www.csbj.org/qbio
#QuantumBiology #ProteinScience #Biophysics #AndersonTransition #Holography #Bioelectronics #Nanotechnology
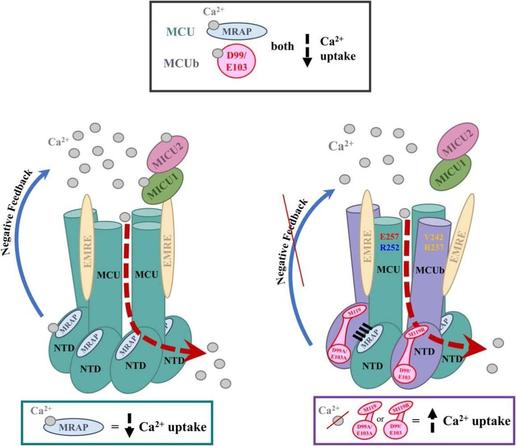
🛠️ Is it possible to rewire cellular energy control by targeting a single protein domain?
🔗 Disrupting the network of co-evolving amino terminal domain residues relieves mitochondrial calcium uptake inhibition by MCUb. Computational and Structural Biotechnology Journal, DOI: https://doi.org/10.1016/j.csbj.2024.12.007
📚 CSBJ: https://www.csbj.org/
#Mitochondria #CellBiology #CalciumSignaling #ProteinEngineering #ProteinScience #StructuralBiology #Bioenergetics #MolecularDynamics #Biophysics #MCU #MCUb #NMR
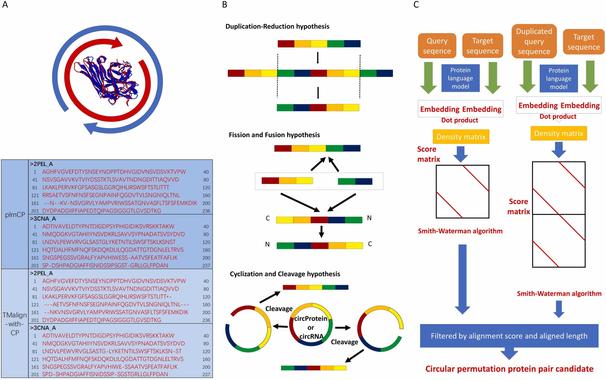
This study introduces plmCP, an innovative method for detecting circular permutations (CPs) in proteins using Protein Language Models (PLMs)—paving the way for deeper insights into protein evolution, engineering, and synthetic biology.
🔗 Detection of circular permutations by Protein Language Models. Computational and Structural Biotech Journal, DOI: https://doi.org/10.1016/j.csbj.2024.12.029
📚 CSBJ: https://www.csbj.org/
#ProteinScience #AIinBiotech #SyntheticBiology #Bioinformatics #ProteinEngineering
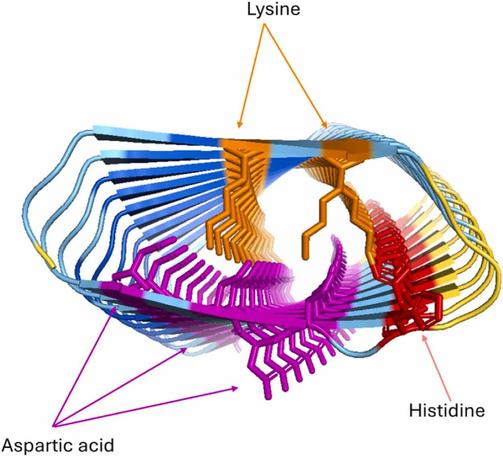
This study investigates the surprising limitations of AlphaFold 2 (AF2) in predicting protein structures—specifically its tendency to confidently predict β-solenoid folds for repeat proteins, even when the structures are unrealistic or unstable.
🔗 AlphaFold 2, but not AlphaFold 3, predicts confident but unrealistic β-solenoid structures for repeat proteins. DOI: https://doi.org/10.1016/j.csbj.2025.01.016
📚 CSBJ: https://www.csbj.org/