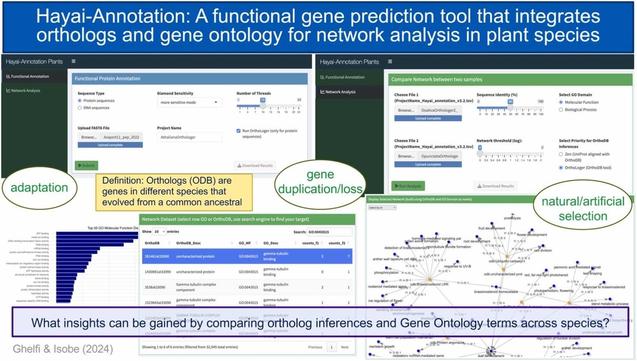
🧬 Can we map the evolutionary history of plant genes through network analysis?
🔗 Hayai-Annotation: A functional gene prediction tool that integrates orthologs and gene ontology for network analysis in plant species. Computational and Structural Biotechnology Journal, DOI: https://doi.org/10.1016/j.csbj.2024.12.011
📚 CSBJ: https://www.csbj.org/
#PlantGenomics #GeneAnnotation #Bioinformatics #Orthologs #RShiny #GeneOntology #ComparativeGenomics #NetworkBiology #Genetics