✍️ New in #eLife: #CellSeg3D introduces #WNet3D, a self-supervised 3D #segmentation method for #microscopy data — no labels needed. Claims to outperform #Cellpose/#StarDist on 4 datasets. Includes #opensource plugin (#Napari) + full 3D annotated #cortex dataset. Will test it later.
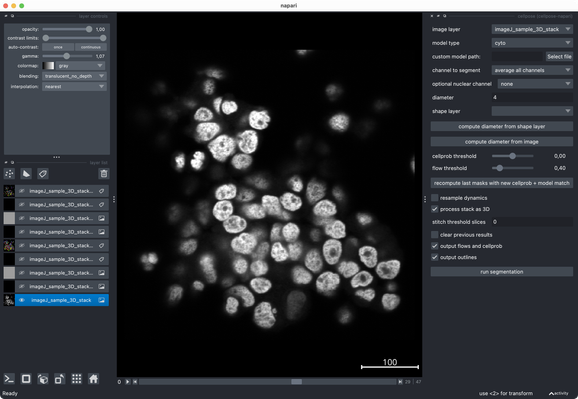
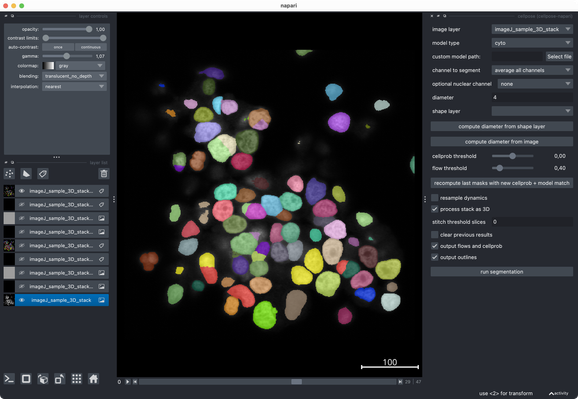
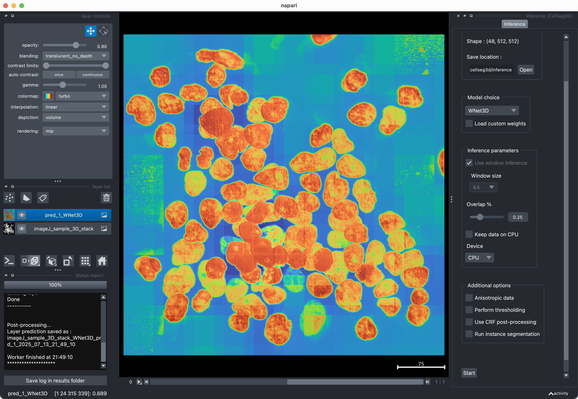
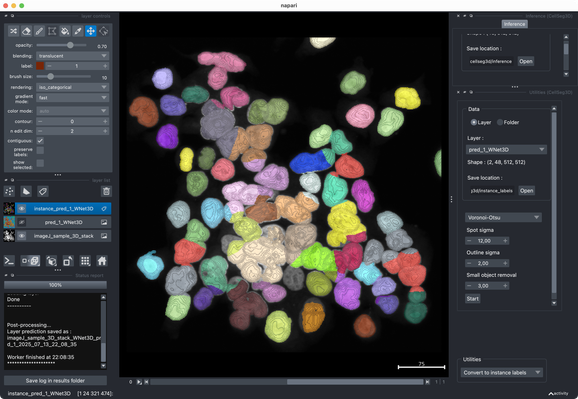
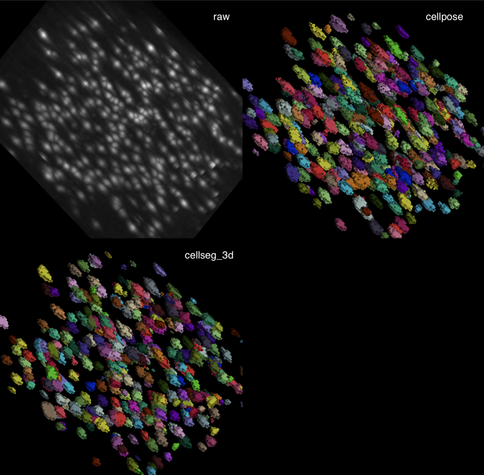
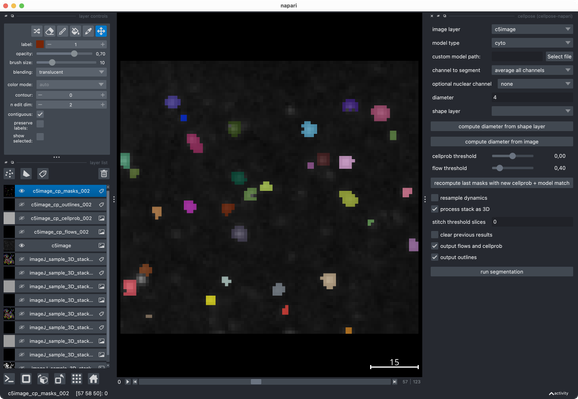
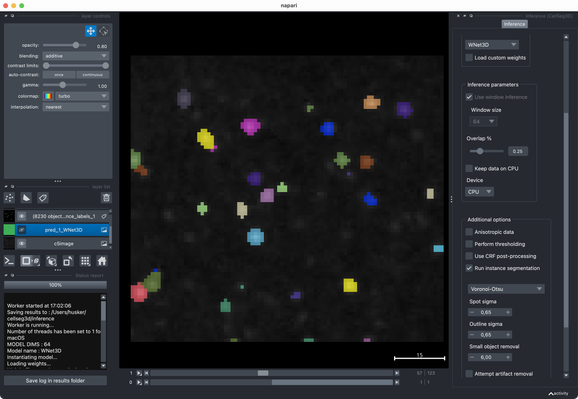
Tried the same with a more realistic 3D stack from the #ImageJ sample library.#Cellpose runs fast and segments very well out of the box.#CellSeg3D takes considerably longer and seems to segment decently, but I couldn’t get a proper instance #segmentation in the post-processing step (which is recommended as part of its workflow). However, #CellSeg3D looks very promising — just needs some more time and parameter exploration, I guess.
I’d recommend giving it a try 👌