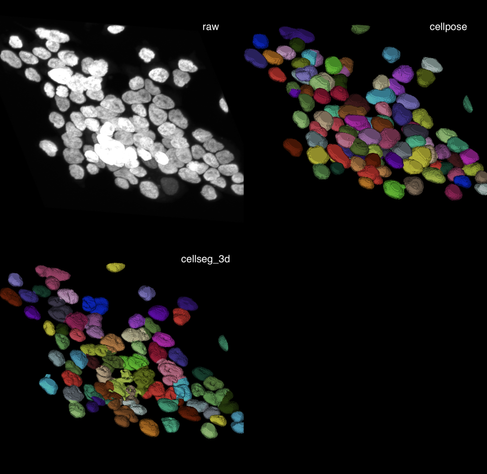
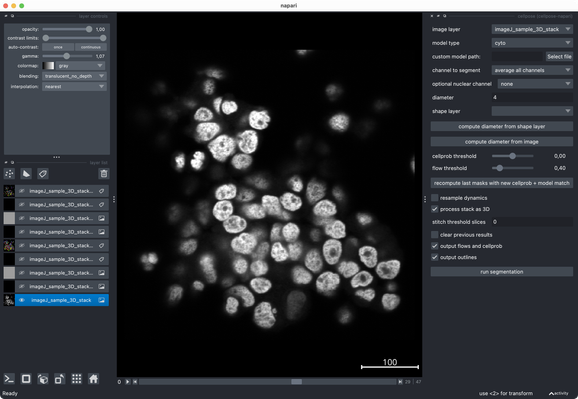
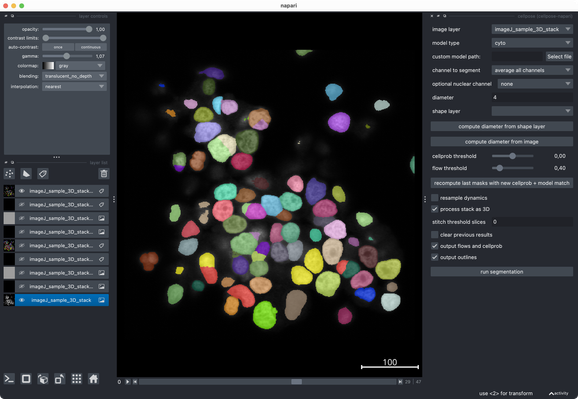
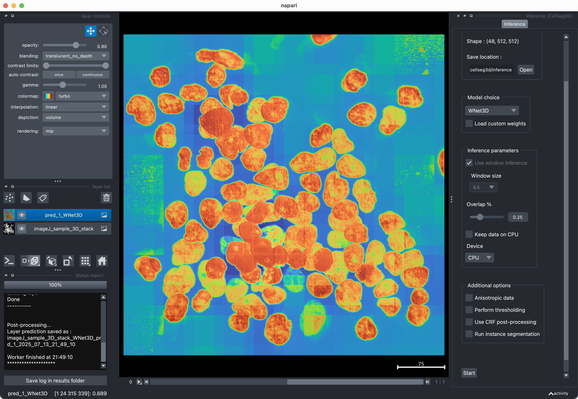
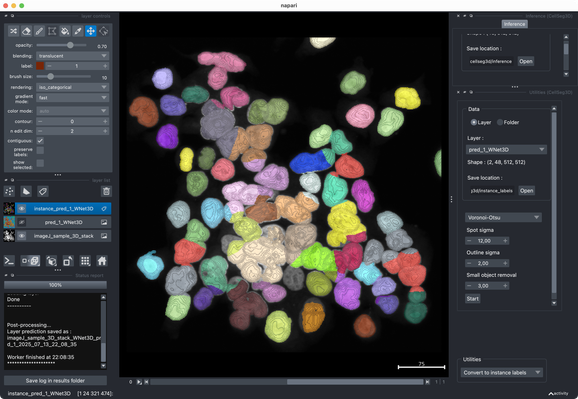
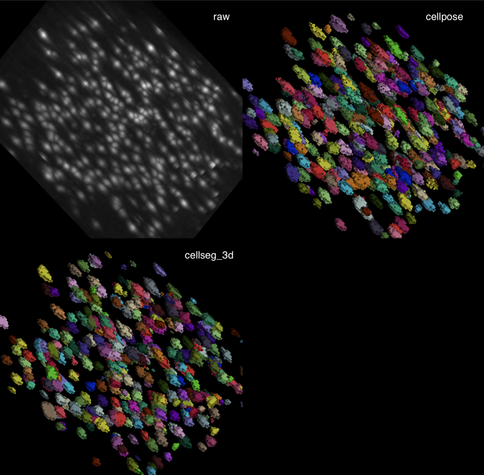
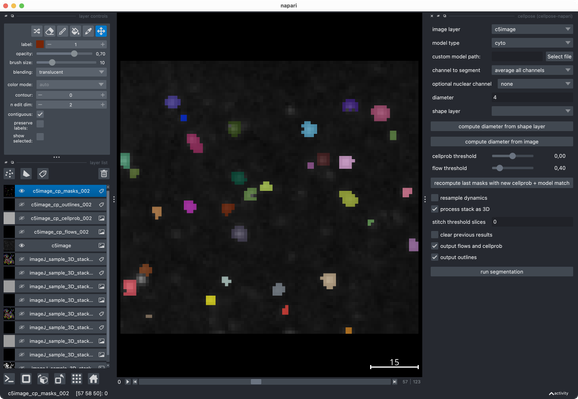
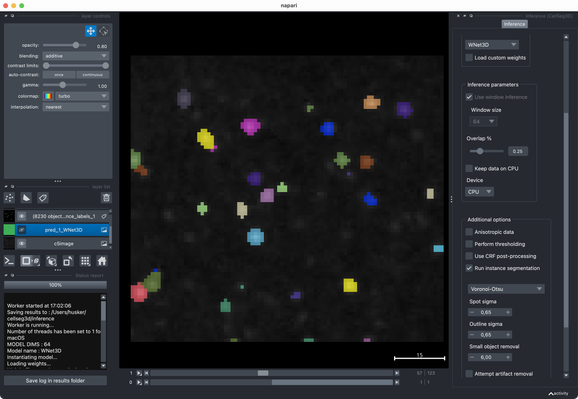
✍️ New in #eLife: #CellSeg3D introduces #WNet3D, a self-supervised 3D #segmentation method for #microscopy data — no labels needed. Claims to outperform #Cellpose/#StarDist on 4 datasets. Includes #opensource plugin (#Napari) + full 3D annotated #cortex dataset. Will test it later.
To wrap this up: Both tools are easy to test. I highly recommend trying them on your own data to see what works best for your use case.
I’ll include #CellSeg3D in our next #Napari #bioimage analysis course (https://www.fabriziomusacchio.com/teaching/teaching_bioimage_analysis/). Curious what impressions and feedback the students will share. 🧪🔍
What I really like about @napari is how well it integrates modern #Python tools. Great to have such a flexible, evolving #opensource platform for (bio) #imageanalysis! 👌