https://doi.org/10.1101/2025.06.09.658708

Multiplets in scRNA-seq data: extent of the problem and efficacy of methods for removal
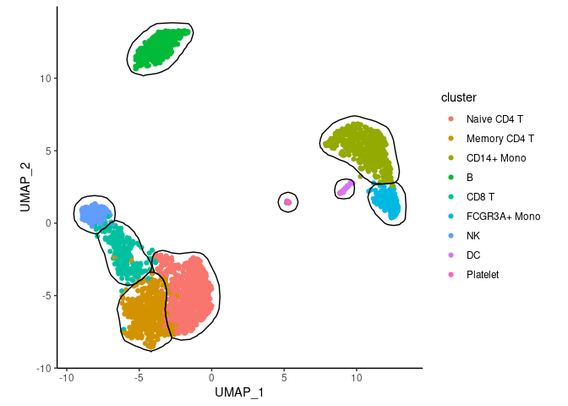
Multiplets-droplets that capture more than one cell-are a known artefact in droplet-based single-cell RNA sequencing (scRNA-seq), yet their prevalence and impact remain underestimated. In this study, we assess the frequency of multiplets across diverse publicly available datasets and evaluate how well commonly used detection tools are able to identify them. Using cell hashing data to determine a lower bound of the true multiplet rate, we demonstrate that commonly used heuristic estimations systematically underestimate multiplet rates, and that existing tools-despite optimized parameters-detect only a small subset of cell-hashing multiplets. We further refine a Poisson-based model to estimate the true multiplet rate, revealing that actual rates can exceed heuristic predictions by more than twofold. Downstream analyses are significantly affected by multiplets: they are not confined to isolated clusters but are distributed throughout the transcriptional landscape, where they distort clustering and cell type annotation. Using both quantitative and qualitative approaches, we visualize these effects and show that cell-hashing-informed multiplet removal eliminates artefactual clusters and improves annotation clarity, whereas computationally detected multiplets fail to fully remove artefacts in the most common experimental contexts. Our findings confirm that multiplet contamination remains a pervasive and under-addressed issue in scRNA-seq analysis. Since most datasets lack multiplexing, researchers must often rely on heuristics and limited tools, leaving many multiplets unidentified. We advocate for more robust multiplet-detection strategies, including multimodal validation, to ensure more accurate and interpretable scRNA-seq results. ### Competing Interest Statement The authors have declared no competing interest. Deutsche Forschungsgemeinschaft, HO 1582/12