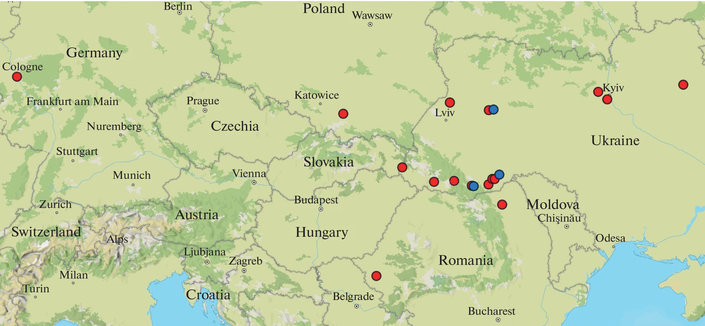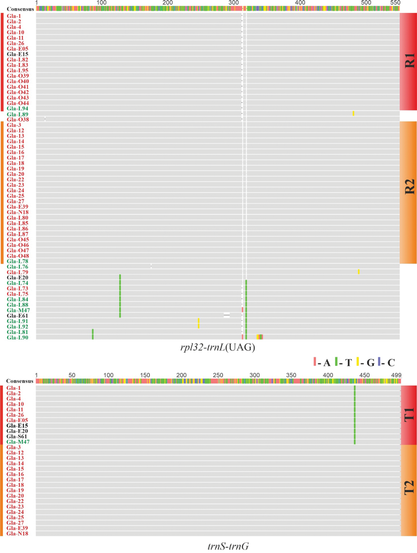
Distribution of Two Chloroplast Haplotypes of the Invasive Weed Himalayan Balsam (Impatiens glandulifera) in Ukraine and other European Countries - Cytology and Genetics
Abstract One of the most well-documented cases of successful plant invasion in Europe is the rapid spread of Himalayan balsam (Impatiens glandulifera Royle). Introduced from the Himalayas to Europe in the first half of the 19th century as an ornamental and nectar-producing plant, this species initially naturalized in the United Kingdom, from where it subsequently dispersed across the European continent. Despite the active invasion of I. glandulifera in Eastern Europe, the genetic diversity of its populations in this region has remained largely unexplored. In this study, we evaluated the variability of two chloroplast DNA (cpDNA) regions, trnS-G and rpl32-trnL(UAG), in I. glandulifera samples from Ukraine and compared the results with cpDNA variants from continental Europe, the United Kingdom, as well as from India and Pakistan. Our results reveal the widespread distribution of two distinct I. glandulifera haplotypes, T1-R1 and T2-R2, across continental Europe. These haplotypes differ in both cpDNA regions analyzed, and their divergence is inferred to have occurred within the species’ native range. Chloroplast DNA variation was found to be significantly higher in the native range than in the invasive range. The broad distribution of the two chloroplast haplotypes across Europe supports the hypothesis of multiple introductions of I. glandulifera into the continent. The uneven distribution of haplotypes T1-R1 and T2-R2 within Ukraine may indicate a founder effect.