New publication out 🆕 📤
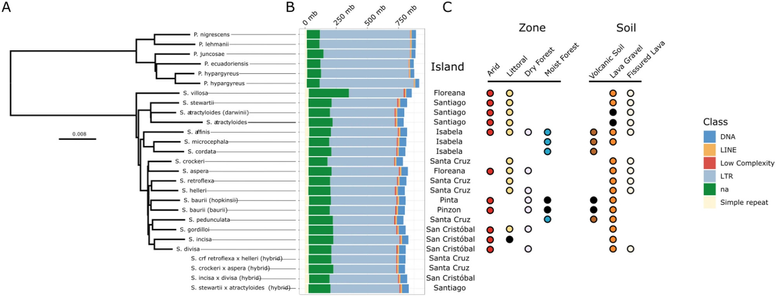
"No evidence of transposable element bursts in the Galápagos Scalesia adaptive radiation despite hybridization, diversification and ecological niche shifts"
https://mobilednajournal.biomedcentral.com/articles/10.1186/s13100-025-00362-z
🔍 Despite rapid speciation, niche shifts, & hybridization, we found no significant TE accumulation differences across species or hybrids.
🌵 Even in arid-adapted lineages—where genome downsizing may be expected—TEs didn’t decline.
#TransposableElements #Speciation #AdaptiveRadiation #Genomics