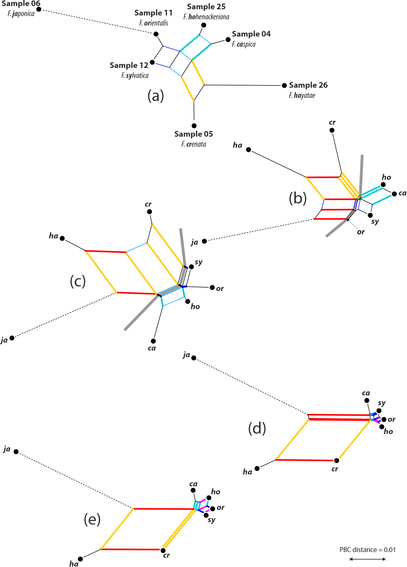
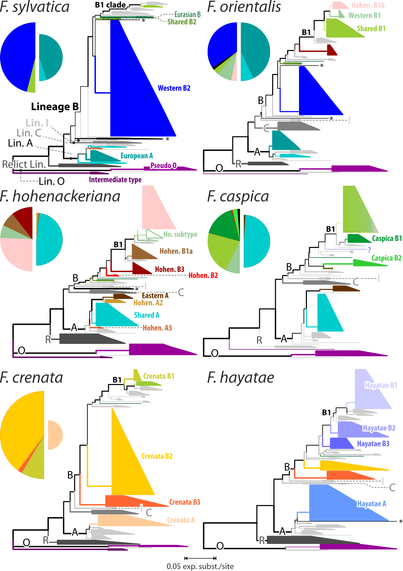
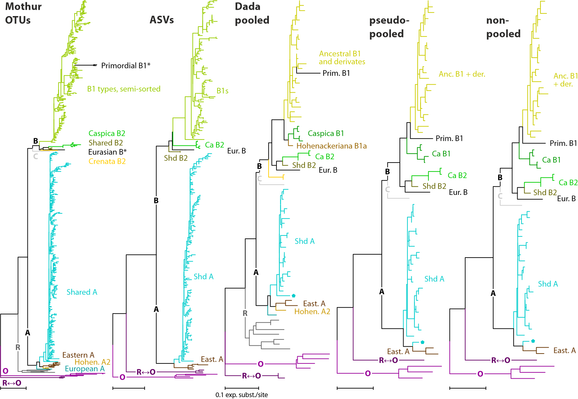
Took a while, but finally out:
Pruning the Tree: Comparing #OTUs and #ASVs in High-Throughput Sequencing of 5S-IGS Nuclear Ribosomal DNA in #Phylogenetic Studies
How to reduced 100ks of reads down to the 100 really needed, a comparison between #Mothur and #dada2 pipelines using our nigh-#reticulate #beech dataset as test subject