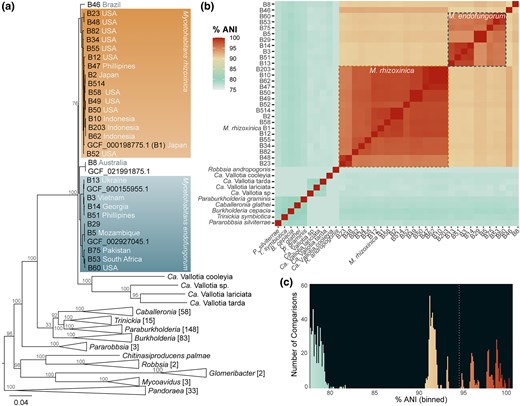
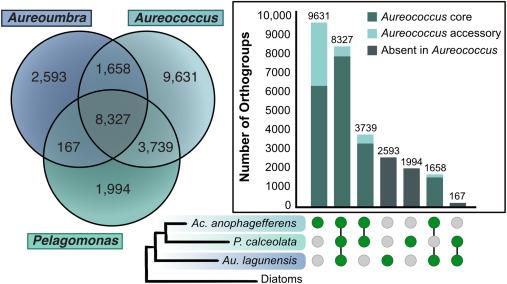
Abbot et al. present a pangenome analysis of the endofungal genus Mycetohabitans, showing bacteria-fungus coevolution.
Going through Samuel Horsfield's presentation on pangenome graphs in comparative genomics, courtesy of EMBL.
Fascinating tidbit on history of alignment based sequence graph - it goes all the way back to 1989 (Hein), era of Apple IIs and IBM PS/2s. Great talk!
Researchers decoded the #pangenome of 33 oat lines, revealing their full #geneticdiversity to support breeding of more resilient, high-yield #crops in the face of #climatechange: http://go.tum.de/651300
📷iStock/ Evgeniy Andreev
I got a shoutout during an EMBL lecture presentation on indexing pangenomes. I have to admit, it made my day 😄
The whole series is fantastic so far, the particular talk on indexing can be found below with slides and video:
https://www.ebi.ac.uk/training/events/making-sense-massive-genomic-data-indexing-scale/
Looks like worth trying
https://www.biorxiv.org/content/10.1101/2025.07.06.663386v1.full.pdf
https://github.com/songbowang125/Swave
#bioinformatics #Genomics #pangenome #ShouldTryThis
Serious #bioinformatics question!
I am testing #pangenome alignment & calling w/ deepvaraint.
The pangenome fasta labels chromosomes with GRCh38#0#chr{0-9}.
While it is annoying that the pangenome ignored chrM, this chr labeling makes it difficult to use any other programs that rely on chr{0-9} for the chr label in the bed or vcf files.
Is it reasonable to relabel the chr using samtools or will it break the pangenome file info?
Delighted to share our latest scientific article (https://doi.org/10.1080/21505594.2025.2504658) about #comparativegenomics of #Salmonella enterica serovars Paratyphi A, Typhi and Typhimurium reveals distinct profiles of their #pangenome, mobile genetic elements, #antimicrobialresistance and defense systems repertoire.
A great collaboration with Charles Coluzzi, Hélène Chiapello and Ohad Gal-Mor !