https://onlinelibrary.wiley.com/doi/10.1002/ece3.71322
https://onlinelibrary.wiley.com/doi/10.1002/ece3.71322
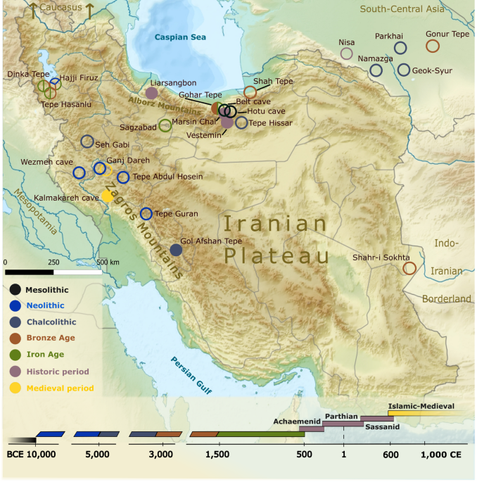
"Ancient DNA indicates 3,000 years of genetic continuity in the Northern Iranian Plateau, from the Copper Age to the Sassanid Empire"
https://www.nature.com/articles/s41598-025-99743-w
#aDNA #ancientDNA #PaleoGenetics #PaleoGenomics #PopulationGenetics
Ancient DNA indicates 3,000 years of genetic continuity in the Northern Iranian Plateau, from the Copper Age to the Sassanid Empire - Scientific Reports
In this study, we present new ancient DNA data from prehistoric and historic populations of the Iranian Plateau. By analysing 50 samples from nine archaeological sites across Iran, we report 23 newly sequenced mitogenomes and 13 nuclear genomes, spanning 4700 BCE to 1300 CE. We integrate an extensive reference sample set of previously published ancient DNA datasets from Western and South-Central Asia, enhancing our understanding of genetic continuity and diversity within ancient Iranian populations. A new Early Chalcolithic sample, predating all other Chalcolithic genomes from Iran, demonstrates mostly Early Neolithic Iranian genetic ancestry. This finding reflects long-term cultural and biological continuity in and around the Zagros area, alongside evidence of some western genetic influence. Our sample selection prioritizes northern Iran, with a particular focus on the Achaemenid, Parthian, and Sassanid periods (355 BCE–460 CE). The genetic profiles of historical samples from this region position them as intermediates on an east-west genetic cline across the Persian Plateau. They also exhibit strong connections to local and South-Central Asian Bronze Age populations, underscoring enduring genetic connections across these regions. Diachronic analyses of uniparental lineages on the Iranian Plateau further highlight population stability from prehistoric to modern times.
🚨 Reminder that the @spaam_community [https://genomic.social/@spaam_community]
will offer its fourth (virtual) summer school course sponsored by the Werner
Siemens Foundation and @NFDI4Microbiota [https://nfdi.social/@NFDI4Microbiota]
this year:
INTRODUCTION TO ANCIENT METAGENOMICS (4-8 Aug 2025)
For more info see https://spaam-community.github.io/wss-summer-school/
[https://spaam-community.github.io/wss-summer-school/] and this 🧵[1/3]
#aDNA [https://genomic.social/tags/aDNA] #Paleogenomics #metagenomics #spaam #AncientMetagenomics #AncientDNA #Microbiome #eDNA
"Mapping the gene regulatory landscape of archaic hominin introgression in modern Papuans"
https://doi.org/10.1101/2025.05.04.652069
(2025-05-04)
#aDNA #ancientDNA #PaleoGenetics #PaleoGenomics #PopulationGenetics

Mapping the gene regulatory landscape of archaic hominin introgression in modern Papuans
Interbreeding between anatomically modern humans and archaic hominins has contributed to the genomes of present-day human populations. However, our understanding of the specific gene regulatory consequences of Neanderthal, and particularly, Denisovan introgression is limited. Here, we used a massively parallel reporter assay to investigate the regulatory effects of 25,869 high-confidence introgressed SNPs segregating in present-day individuals of Papuan genetic ancestry in immune cell types. Overall, 8.22% of Denisovan and 8.58% of Neanderthal sequences showed active regulatory activity, and 9.22% of these displayed differential activity between alleles. We examined the properties of active and differentially active sequences, and found no impact of introgressed allele frequency on the probability of activity, but do observe an association with distance to the nearest transcription start site. Differential activity was also associated with differential transcription factor binding. Genes predicted to be regulated by differentially active sequences included IFIH1 and TNFAIP3, key immune genes and known examples of archaic introgression. Overall, this work provides insight into the regulatory activity of archaic variants in Papuan populations and, more broadly, the contribution of archaic introgression in shaping modern human genetic diversity. ### Competing Interest Statement The authors have declared no competing interest. Leakey FoundationLeakey Foundation, https://ror.org/018kdpd27, Australian Research CouncilAustralian Research Council, https://ror.org/05mmh0f86, DP200101552 Royal Society of New ZealandRoyal Society of New Zealand, https://ror.org/04tajb587, 20-MAU-017 Agence Nationale de la RechercheAgence Nationale de la Recherche, , PAPUAEVOL ANR-20CE12-0003-01 Estonian Research CouncilEstonian Research Council, , TK214
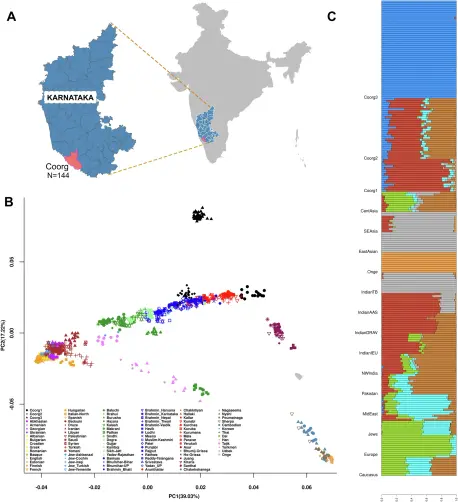
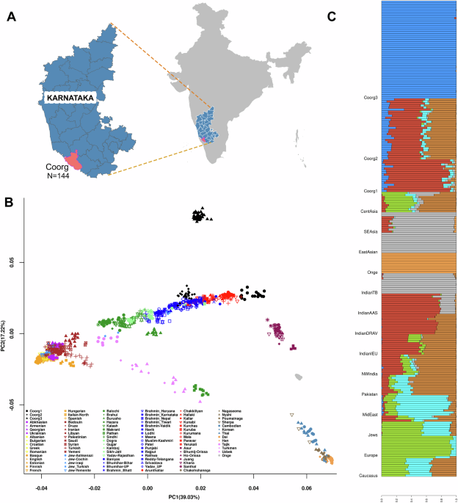
"Unique demographic history and population substructure among the Coorgs of Southern India"
https://www.nature.com/articles/s42003-025-08073-0
(2025-05-05)
#aDNA #ancientDNA #PaleoGenetics #PaleoGenomics #PopulationGenetics
Unique demographic history and population substructure among the Coorgs of Southern India - Communications Biology
Coorgs from southwestern India are a culturally homogeneous but genetically heterogeneous group and earliest descendants of the Bronze Age migration into South Asia with enhanced ancestry proportions of the Middle East and population-specific drift.
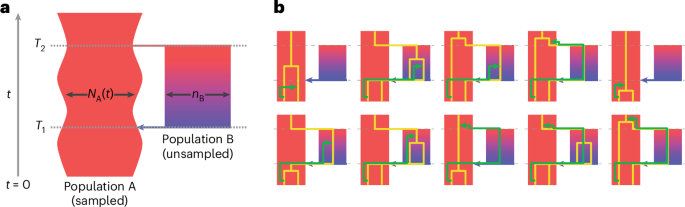
"A structured coalescent model reveals deep ancestral structure shared by all modern humans"
https://www.nature.com/articles/s41588-025-02117-1
(2025-03-18)
#aDNA #ancientDNA #PaleoGenetics #PaleoGenomics #PopulationGenetics
A structured coalescent model reveals deep ancestral structure shared by all modern humans - Nature Genetics
The cobraa model extends the pairwise sequentially Markovian coalescent to identify structured population history by examination of the model transition matrix. Applied to human polymorphism data, cobraa identifies an ancient admixture event ancestral to all modern humans.
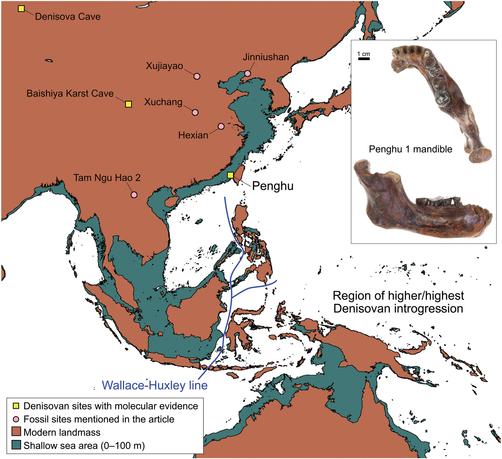
"A male Denisovan mandible from Pleistocene Taiwan"
https://doi.org/10.1126/science.ads3888
(2025-04-10)
"Does Neanderthal DNA put a ceiling on [the] Out of Africa [event]"
by Stefan Milo
(2025-04-10)

Does Neanderthal DNA put a ceiling on Out of Africa?
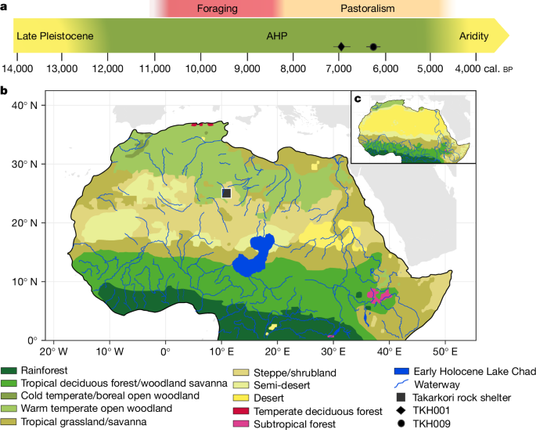
"Ancient DNA from the Green Sahara reveals ancestral North African lineage"
https://www.nature.com/articles/s41586-025-08793-7
(2025-04-02)
#aDNA #PaleoGenetics #PaleoGenomics #PopulationGenetics #Africa #NorthAfrica #GreenSahara
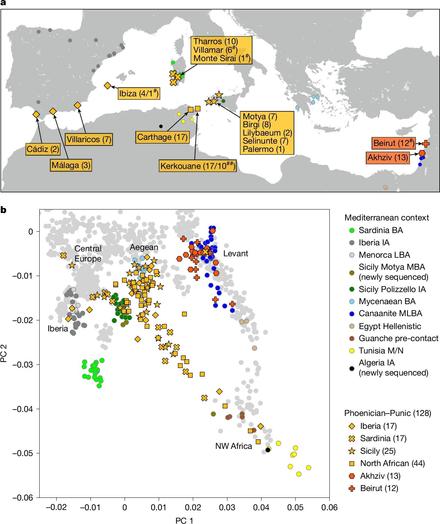
"Punic people were genetically diverse with almost no Levantine ancestors"
https://www.nature.com/articles/s41586-025-08913-3
(2025-04-23)
...
Comment from Iosif Lazaridis:
"""
Consider this surprising fact:
There was a time during the 1st millennium BC that
a. in the West Mediterranean, Greek descendants advanced the Punic cause, and;
b. in the Levant, Phoenician descendants advanced Hellenism.
"""
#aDNA #PaleoGenetics #PaleoGenomics #PopulationGenetics #Greek #Punic
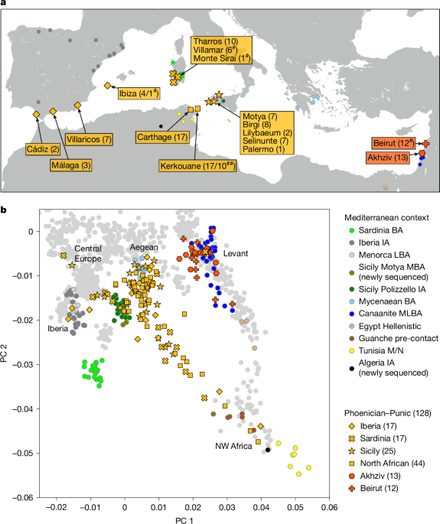
Punic people were genetically diverse with almost no Levantine ancestors - Nature
Levantine Phoenicians made little genetic contribution to Punic settlements in the central and western Mediterranean between the sixth and second centuries bce; instead, the Punic people derived most of their ancestry from a genetic profile similar to that of Sicily and the Aegean, with notable contributions from North Africa as well.