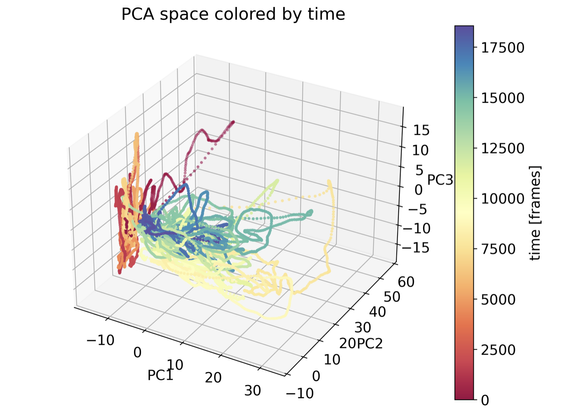
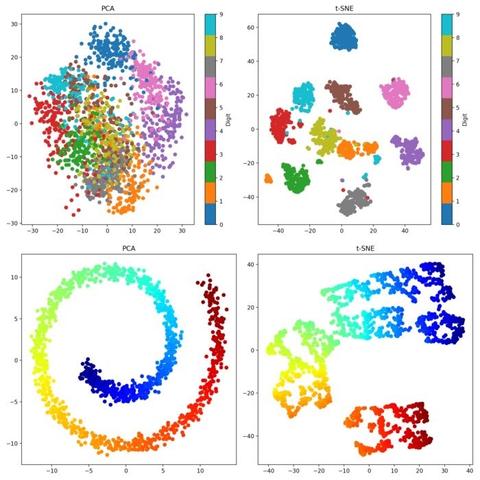
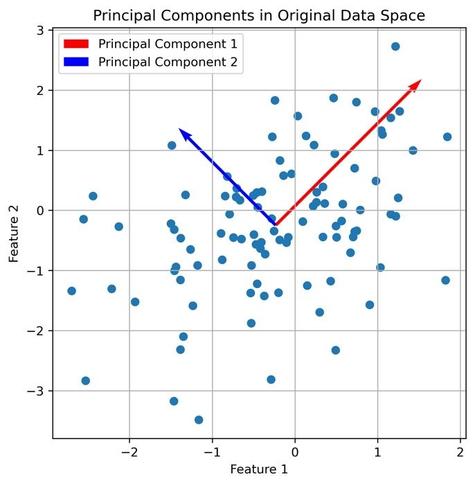
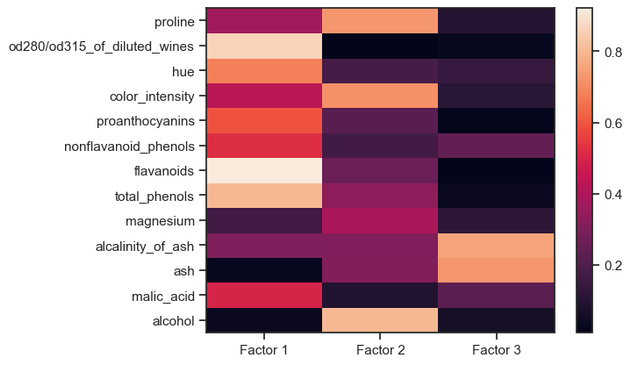
We are excited to welcome Prof. Alejandro Rodriguez Garcia from the Abdus Salam International Centre for Theoretical Physics (ICTP) to Enabla! In his lecture, Alex continues the topic started by Marcello and explores the use of unsupervised machine learning techniques in many-body quantum systems, highlighting how dimensionality reduction can illuminate structure within complex data. Particular emphasis is placed on the Principal Component Analysis (PCA) as a key method to maximize variance while reducing dimensionality. This lecture sets the stage for future topics such as clustering and manifold learning.
🎥 Join us for this #OpenAccess lecture and take advantage of Enabla's unique features to ask questions directly to Prof. Rodriguez Garcia and engage in discussions with the community: https://enabla.com/pub/1112/about
Don't miss this opportunity to enhance your knowledge in the intersection of data mining and quantum physics!
#MachineLearning #UnsupervisedLearning #DimensionalityReduction #QuantumSystems #PCA #DataMining #OpenScience