🔍 Can one hundred scans be linked to hearing loss? The case of the Courtois NeuroMod project
For over five years, the Courtois NeuroMod project scanned six participants weekly using fMRI — creating the largest individual-subject fMRI dataset ever collected.
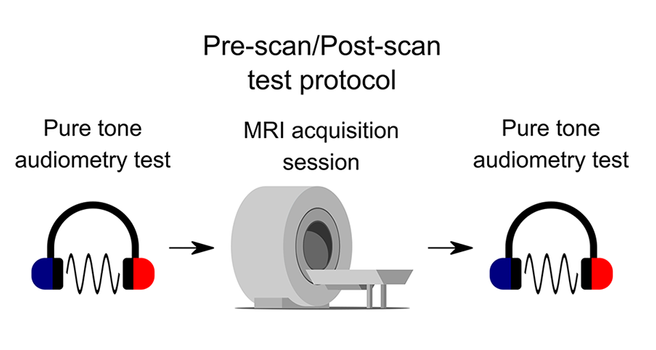
MRI machines are loud, and participants wore MRI-compatible Sensimetrics earphones with foam inserts and additional custom over-the-ear protection. We still remained vigilant about potential impacts on auditory health.
📣 A new study, led by Eddie Fortier under the supervision of Adrian Fuente, and now published in PLOS ONE, presents the results of an auditory monitoring protocol conducted in parallel with CNeuroMod:
🔗 https://lnkd.in/eRinvsDH
Key Findings:
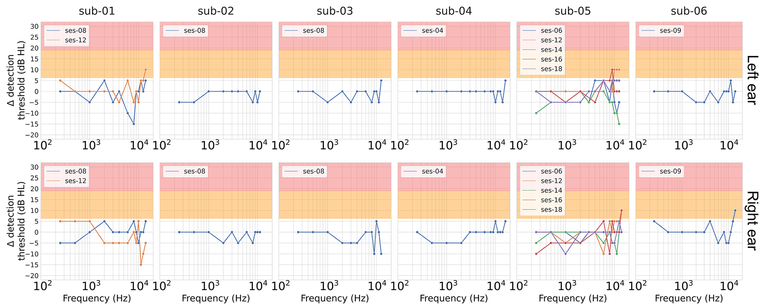
Across participants, we found no clinical signs of ear trauma immediately following scanning. Changes in detection thresholds were typically <10 dB, even in high-frequency ranges (>10 kHz) where variability was greatest.
One participant with pre-existing unilateral hearing loss was tested across five sessions. Their results were inconsistent — and in some cases, paradoxically showed improved sensitivity post-scan — likely due to test-retest variability and fatigue effects in the upper frequency range.
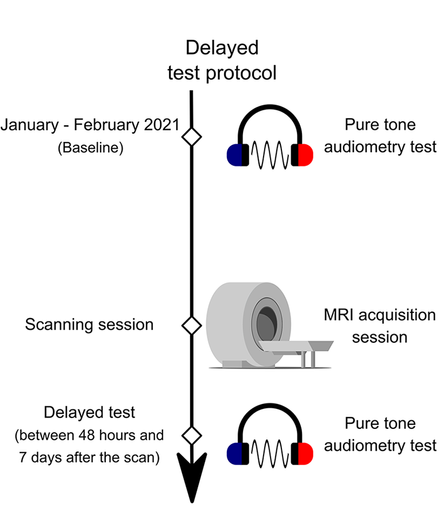
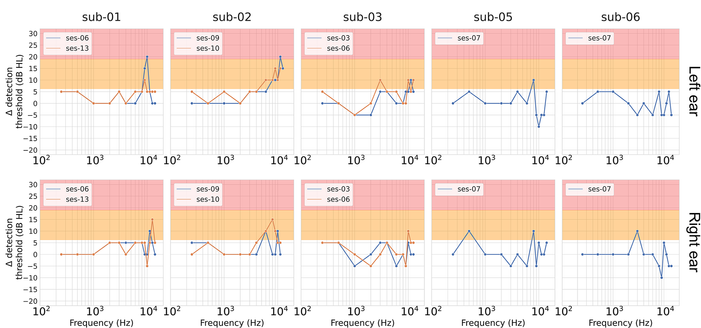
In long-term follow-up (up to 16 months delay), we observed no sustained hearing loss. While high-frequency measures remained variable, no clinically significant, consistent declines were found across the group.
🎧 While pure tone audiometry is a cognitively demanding test — especially following extended scanning sessions — our findings are reassuring: with proper hearing protection, even repeated, long-duration fMRI protocols like CNeuroMod can be conducted safely. See the paper for full results and a complete discussion.
The five-year CNeuroMod data collection phase is now complete, and we are deeply grateful to the participants who committed their time to this study, and to the Courtois Foundation for their visionary support.
We are now preparing a series of public data releases and publications that will continue to explore the many facets of this unique longitudinal dataset.
Stay tuned.