Robert Haase
@haesleinhuepf
- 224 Followers
- 81 Following
- 39 Posts
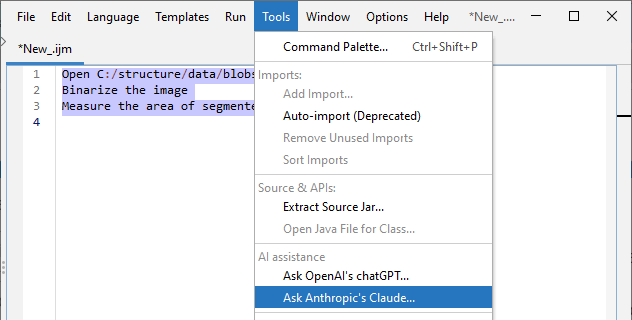
I made a new version of @FijiSc's Script Editor that supports #AIassisted macro coding using @AnthropicAI's #claude and @OpenAI's #GPT. Read more about how you can try out: https://forum.image.sc/t/ai-assisted-code-generation-in-fijis-script-editor/100877?u=haesleinhuepf
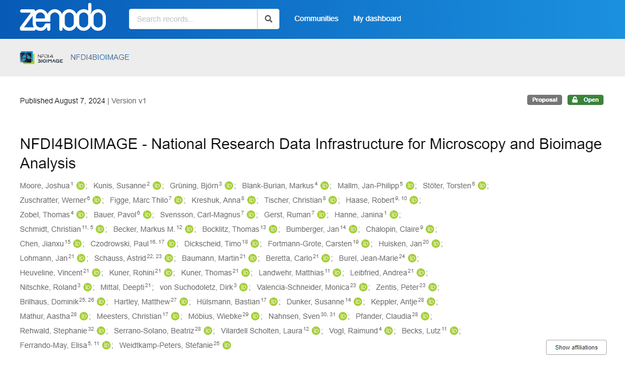
Looking for some summer holiday's literature? @nfdi4bioimage has now published its 2021 grant proposal to take part in the @NFDI https://doi.org/10.5281/zenodo.13168693
Read about our vision, strategy, task areas & measures.
We look forward to increasing interactions to facilitate FAIR bioimaging data.
Read about our vision, strategy, task areas & measures.
We look forward to increasing interactions to facilitate FAIR bioimaging data.
NFDI4BIOIMAGE - National Research Data Infrastructure for Microscopy and Bioimage Analysis
Bioimaging refers to a collection of methods to visualize the internal structures and mechanisms of living organisms. The fundamental tool, the microscope, has enabled seminal discoveries like that of the cell as the smallest unit of life, and continues to expand our understanding of biological processes. Today, we can follow the interaction of single molecules within nanoseconds in a living cell, and the development of complete small organisms like fish and flies over several days starting from the fertilized egg. Each image pixel encodes multiple spatiotemporal and spectral dimensions, compounding the massive volume and complexity of bioimage data. Proper handling of this data is indispensable for analysis and its lack has become a growing hindrance for the many disciplines of the life and biomedical sciences relying on bioimaging. No single domain has the expertise to tackle this bottleneck alone. As a method-specific consortium, NFDI4BIOMAGE seeks to address these issues, enabling bioimaging data to be shared and re-used like they are acquired, i.e., independently of disciplinary boundaries. We will provide solutions for exploiting the full information content of bioimage data and enable new discoveries through sharing and re-analysis. Our RDM strategy is based on a robust needs analysis that derives not only from a community survey but also from over a decade of experience in German BioImaging, the German Society for Microscopy and Image Analysis. It considers the entire lifecycle of bioimaging data, from acquisition to archiving, including analysis and enabling re-use. A foundational element of this strategy is the definition of a common, cloud-compatible, and interoperable digital object that bundles binary images with their descriptive and provenance metadata. With members from plant biology to neuroscience, NFDI4BIOIMAGE will champion the standardization of bioimage data to create a framework that answers discipline-specific needs while ensuring communication and interoperability with data types and RDM systems across domains. Integration of bioimage data with, e.g., omics data as the basis for spatial omics, holds great promise for fields such as cancer medicine. Unlocking the full potential of bioimage data will rely on the development and broad availability of exceptional analysis tools and training sets. NFDI4BIOIMAGE will make these accessible and usable including cutting-edge AI-based methods in scalable cloud environments. NFDI4BIOIMAGE intersects with multiple NFDI consortia, most prominently with GHGA for linking image and genomics data and with DataPLANT on the definition of FAIR data objects. Last but not least, NFDI4BIOIMAGE is internationally well connected and represents the opportunity for German scientists to keep path with and have a voice in several international initiatives focusing on the FAIRification of bioimage data as one of the main challenges for the advancement of knowledge in the life and biomedical sciences.
Creative Commons itself takes the, IMO very reasonable, view that training AI models on copyrighted works constitutes fair use:
https://creativecommons.org/2023/08/18/understanding-cc-licenses-and-generative-ai/
The folks who are calling such training "theft" might regret what they seem to be implicitly asking for, i.e. much stricter copyright. Copyright law won't prevent Microsoft, Google, OpenAI or Adobe from making shady licensing deals, but they'll prevent the free/open community from keeping up.

Understanding CC Licenses and Generative AI - Creative Commons
Many wonder what role CC licenses, and CC as an organization, can and should play in the future of generative AI. The legal and ethical uncertainty over using copyrighted inputs for training, the uncertainty over the legal status and best practices around works produced by generative AI, and the implications for this technology on the…
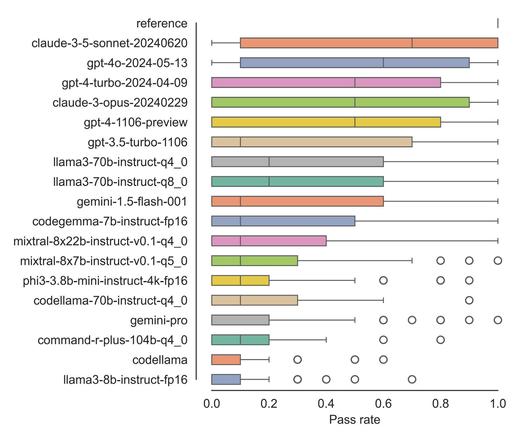
#preprint alert 🚨
Can Large Language Models solve common #BioImageAnalysis coding tasks? And which #LLM performs best? 🙃🤓
Great collaboration with Christian Tischer, Jean-Karim Hériché and Nico Scherf
🔬🖥️🅰️ℹ️🤝🚀
https://www.biorxiv.org/content/10.1101/2024.04.19.590278v3
Can Large Language Models solve common #BioImageAnalysis coding tasks? And which #LLM performs best? 🙃🤓
Great collaboration with Christian Tischer, Jean-Karim Hériché and Nico Scherf
🔬🖥️🅰️ℹ️🤝🚀
https://www.biorxiv.org/content/10.1101/2024.04.19.590278v3
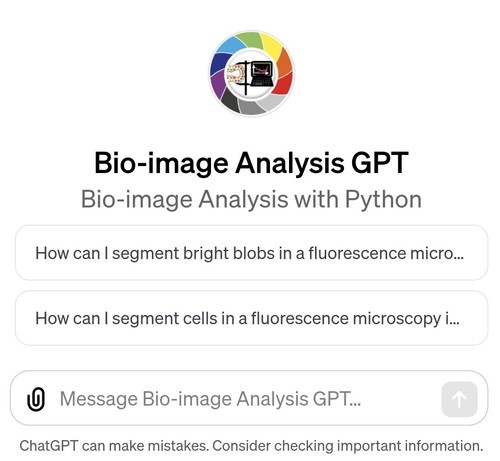
I made a #BioImageAnalysis #GPT 🎉
Compared to #ChatGPT it provides more concise hints and knows more about some Python libraries my collaborators and yours truly maintain. Thanks to all contributors and as always: Feedback is 💚ly welcome 🤗
https://chat.openai.com/g/g-psAohb1OY-bio-image-analysis-gpt
Over the moon announcing: Effectively today I joined the Data Science Center https://ScaDS.AI at Uni Leipzig as Bio-image Data Scientist, Lecturer and Training Coordinator 🔬🖥️🚀
Thanks for the warm welcome everyone 🤗 Exciting days ahead 😃
Thanks for the warm welcome everyone 🤗 Exciting days ahead 😃
Teaching academic career paths to cats, lesson 212: Signing a permanent contract
#IchBinHanna
#IchBinHanna
PhD fellowship available in our team: How does regeneration differ from development? Explore how animals regenerate and to what extent this process mirrors/recapitulates what happens during embryonic development
Host lab in Lyon, France: https://www.averof-lab.org/
PhD student will be part of the REGENERATE-IT Marie Curie network (https://www.regenerate-it.eu), including training, research exchanges in labs across Europe, being part of exciting network of young researchers. International environment and attractive 3-year salary.
Working language in the team/network is English (candidates do not need to speak french, as implied in the ad)
More details here: https://euraxess.ec.europa.eu/jobs/71013
Apply here: https://emploi.cnrs.fr/Offres/Doctorant/UMR5242-MICAVE-012/Default.aspx?lang=EN
RT @haesleinhuepf
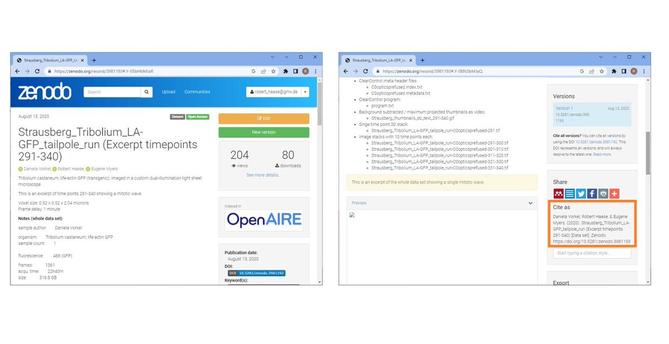
Sharing research data #openaccess is not complicated😌
I just published a @focalplane_jcs blog post about how to share data #FAIR-ly with @ZENODO_ORG
🔬🖥️➡️🌎 #lovedata2023 #LoveDataWeek
https://focalplane.biologists.com/2023/02/15/sharing-research-data-with-zenodo/
As usual: Feedback is ❤️ly welcome!
Sharing research data #openaccess is not complicated😌
I just published a @focalplane_jcs blog post about how to share data #FAIR-ly with @ZENODO_ORG
🔬🖥️➡️🌎 #lovedata2023 #LoveDataWeek
https://focalplane.biologists.com/2023/02/15/sharing-research-data-with-zenodo/
As usual: Feedback is ❤️ly welcome!