[Preprint alert]
We’re thrilled to introduce EnrichMap: a powerful method for spatial gene set enrichment. It’s spatially aware, biologically meaningful and built for the next generation of spatial transcriptomics.
https://www.biorxiv.org/content/10.1101/2025.05.30.656960v1Spatial transcriptomics is transforming how we see tissues but traditional enrichment ignores space. EnrichMap fixes that. We bring spatial context into the heart of gene set scoring.
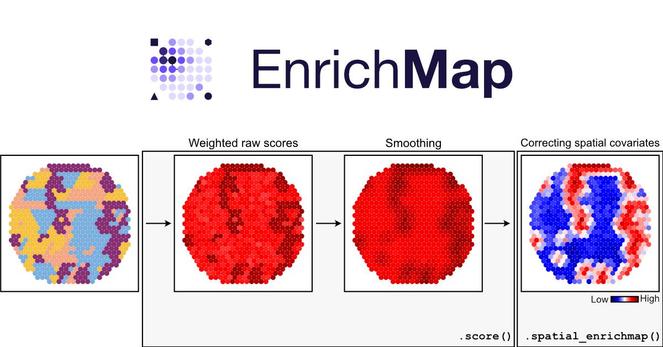
EnrichMap doesn’t just average gene sets over spots. It builds a spatial graph, smooths expression, and captures neighbourhood-level biology. You see what’s really happening across tissue.
Why does this matter?
Tumours are messy. Immune niches are local. Cell states vary in space. If you’re collapsing expression without space, you’re missing the story. EnrichMap keeps that story intact.
What can EnrichMap do?
- Correct for spatial confounders
- Support batch correction and gene weighting
- Map gradients of tumour and immune programmes
- Compare microenvironments
- Reveal niche-specific signalling
We’ve designed it to be fast, flexible and easy to use:
- Works seamlessly with Scanpy, Squidpy and AnnData (scverse compatible)
- Plug-and-play any ST dataset
- Outputs spatially informed scores
- Geostatistics-inspired metrics to evaluate gene sets