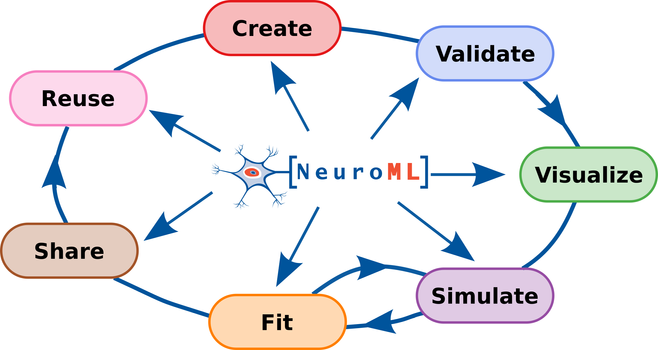
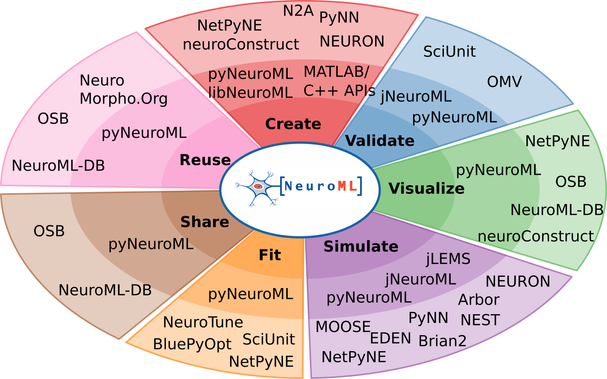
We are very happy to provide a consolidated update on the #NeuroML ecosystem in our @eLife paper, “The NeuroML ecosystem for standardized multi-scale modeling in neuroscience”: https://doi.org/10.7554/eLife.95135.3
#NeuroML is a standard and software ecosystem for data-driven biophysically detailed #ComputationalModelling endorsed by the @INCF and CoMBINE, and includes a large community of users and software developers.
#Neuroscience #ComputationalNeuroscience #ComputationalModelling 1/x