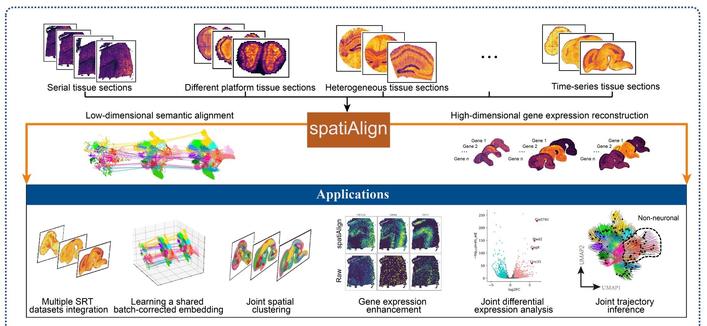
Latest addition to our #SpatialOmics Methods series tackles the challenge of integrating multiple spatially resolved transcriptomics datasets. That employs the expression of all measured genes and the spatial location of cells, to integrate multiple tissue sections.
spatiAlign: an unsupervised contrastive learning model for data integration of spatially resolved transcriptomics https://doi.org/10.1093/gigascience/giae042
See the series here: https://academic.oup.com/gigascience/pages/spatial-omics-methods-and-applications